[21.02] TransUNet

Convolutions Are Not Enough
TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation
ViT was proposed at the end of 2020, and its applications quickly expanded across many fields.
Here, we examine a paper that combines U-Net and ViT. The paper itself isn't difficult; as long as you're familiar with the concepts of U-Net and ViT, you'll be able to grasp it easily.
For those who need a refresher, you can refer to our previous articles:
Defining the Problem
Introducing attention mechanisms into CNNs isn't a new concept, but applying it to medical image segmentation remains rare.
No one has done it? Well, that's the biggest problem!
The authors wanted to use a "sequence-to-sequence" approach, much like ViT, by splitting images into many patches and feeding them directly into a Transformer, learning the global features of the image in the process.
However, if we simply use upsampling to resize the low-level features produced by the Transformer to the original size (e.g., using Upsample), we would only get coarse segmentation results. While Transformer excels at processing global information, it doesn't inherently enhance local boundaries and details.
In ViT, the patch size is usually set at 16×16, which is sufficient for image classification. But for segmentation tasks, we need precision down to the pixel level, which means reducing the patch size to 8×8 or even 4×4. However, the smaller the patch size, the higher the computational cost, which would result in Transformer calculations exploding and making training impossible.
So, we need to first address the issues of "multi-scale downsampling" and "resolution."
Solving the Problem
Clearly, directly applying ViT to image segmentation tasks is not feasible, so the authors propose a hybrid architecture, as shown in the diagram below:
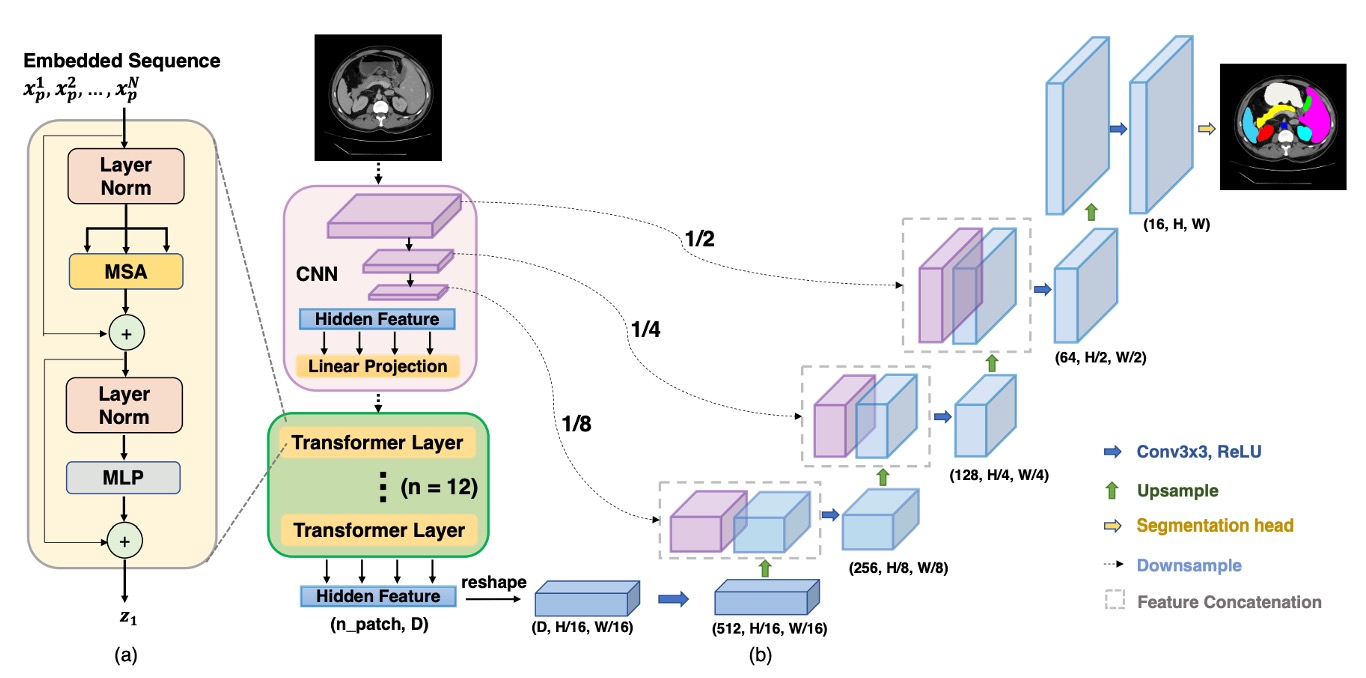
First, CNN is used for feature extraction, like a typical Backbone, which generates feature maps at 1/2, 1/4, 1/8, and 1/16 resolutions.
Given the computational complexity of Transformer, only the 1/16 resolution feature maps are used, paired with a patch size, converting the image into a sequence, which is then passed through a Transformer for global feature extraction.
After the Transformer performs self-attention operations, it produces a more "global" feature representation, which is then transformed back into a 1/16 resolution feature map. The CNN's progressive upsampling (like U-Net's decoder) is used to fuse feature maps at other resolutions. In this way, we retain the advantages of global information while using skip connections to supplement local details, resulting in finer segmentation.
We omit the detailed implementation of ViT and U-Net here, as we have covered them before.
Discussion
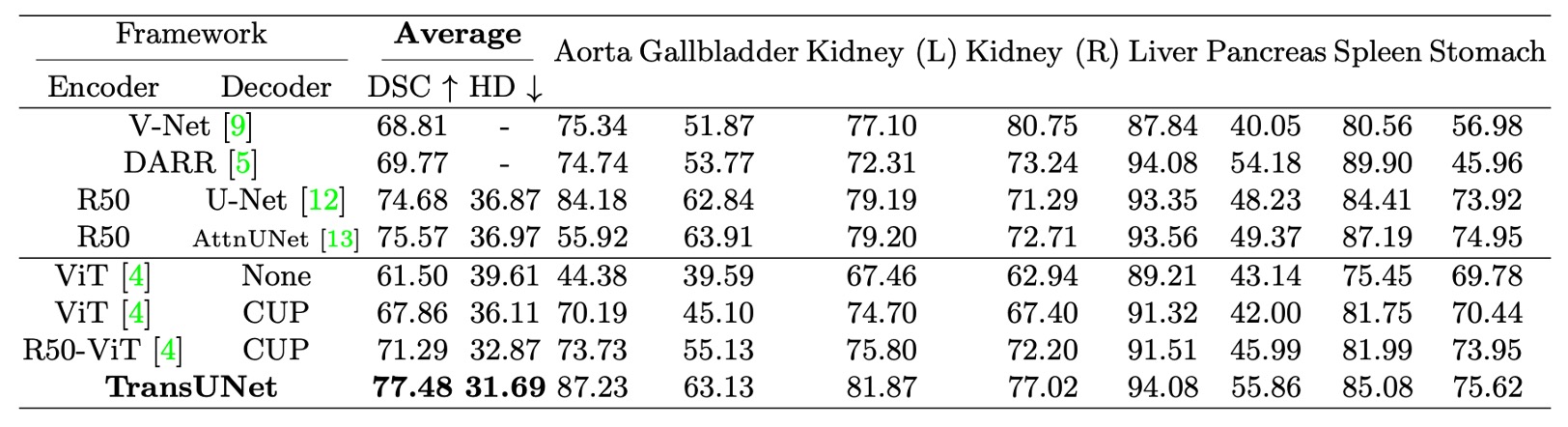
- The table above shows the use of Cascaded Upsampler (CUP) to compare the performance of direct upsampling.
On the Synapse Multi-organ Segmentation Dataset, the authors compared TransUNet with four previous SOTA methods: V-Net, DARR, U-Net, and AttnUNet.
Additionally, to validate the effectiveness of CUP and Hybrid Encoder, the experiments compared the following variants:
- ViT-None: ViT as the encoder, using simple upsampling for decoding
- ViT-CUP: ViT as the encoder, paired with CUP decoder
- R50-ViT-CUP: ResNet-50 + ViT as the encoder, paired with CUP decoder
- TransUNet: R50-ViT-CUP + U-Net style skip connections
To ensure a fair comparison, the encoders of U-Net and AttnUNet were also replaced with ImageNet-pretrained ResNet-50, aligning them with the ViT-Hybrid version.
- Direct Upsampling vs. CUP: Comparing ViT-None vs. ViT-CUP, the Dice Similarity Coefficient (DSC) improved by 6.36%, and the Hausdorff distance decreased by 3.50 mm. This demonstrates that the CUP decoder is better than simple upsampling for medical image segmentation, as it restores boundary details more precisely.
- ViT vs. R50-ViT: Comparing ViT-CUP vs. R50-ViT-CUP, DSC improved by an additional 3.43%, and Hausdorff distance decreased further by 3.24 mm. The conclusion is that while pure ViT captures high-level semantics, it struggles to preserve the boundaries and details of medical images. The hybrid CNN + ViT encoder effectively compensates for the loss of low-level features.
Finally, comparing TransUNet to other methods, DSC increased by 1.91% to 8.67%. Compared to the best CNN method, R50-AttnUNet, TransUNet showed an additional improvement of 1.91%. Compared to R50-ViT-CUP, TransUNet improved by 6.19%.
The authors explain that pure CNNs, while good at capturing rich local details, lack a global perspective. Pure ViT, while effective at capturing global semantics, struggles to retain boundaries and details. TransUNet, by combining global and local information through skip connections, successfully surpasses all SOTA methods, becoming the new benchmark for medical image segmentation.
Conclusion
TransUNet leverages the powerful global self-attention mechanism of Transformers combined with the low-level detail representation of CNNs, overcoming the limitations of traditional FCN architectures and demonstrating exceptional performance in medical image segmentation.
Through its U-shaped hybrid architecture, TransUNet outperforms existing CNN-based self-attention methods, providing a novel solution for medical image segmentation and proving the potential and practicality of Transformers in this field.